Background. Coagulopathic bleeding is a major cause of mortality after trauma, and platelet dysfunction contributes to this problem. The causes of platelet dysfunction are unknown, but a great deal can be learned from the plasma environment after injury, which may directly alter platelet function. Studying the changes in plasma using untargeted proteomics would provide unbiased insight into the presence of possible inhibitors of platelet function, changes to their major ligands, or other previously unknown pathways affecting platelet function.
Methods. Citrated blood was collected from severely injured trauma patients at the time of their arrival to the Emergency Department. Platelet testing was performed immediately, and plasma was frozen for analysis. Samples were collected from 110 patients, and a subset of 24 patients was identified by a preserved (n=12) or severely impaired (n=12) platelet aggregation response to five different agonists (adenosine diphosphate, arachidonic acid, collagen, thrombin receptor-activating peptide, and ristocetin). Untargeted proteomics was performed by nanoflow liquid chromatography tandem mass spectrometry to determine the plasma protein profile associated with platelet dysfunction. Protein abundance levels for each patient were normalized to total protein concentration to control for hemodilution by crystalloid fluid infusion prior to blood draw. Results were compared by Wilcoxon rank-sum test, and a two-tailed p value less than 0.05 was considered significant. No adjustment was made for multiple comparisons, as the risk of a type II error was felt to outweigh that of a type I error in this exploratory analysis.
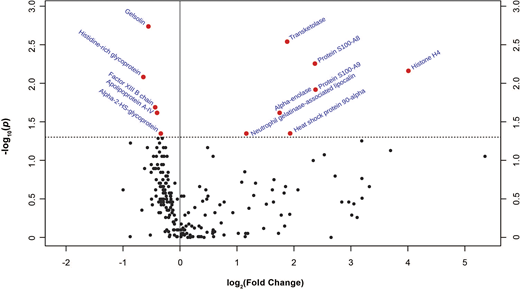
Results. Patients with platelet dysfunction were more severely injured (median Injury Severity Score 29.5 vs. 13.5, p=0.002) but otherwise demographically similar to those with retained platelet function. Of 232 proteins identified, twelve were significantly different in the low- vs. high-platelet function groups: gelsolin (median peak intensity 9.70E+6 vs. 1.48 E+7, p=0.002), transketolase (2.86E+4 vs. 9.36E+3, p=0.003), protein S100-A8 (4.60E+4 vs. 2.64E+4, p=0.006) and -A9 (5.83E+4 vs. 4.07E+4, p=0.012), histone H4 (5.72E+4 vs. 8.47E+6, p=0.007), histidine-rich glycoprotein (HRG) (5.00E+6 vs. 8.30E+6, p=0.008), factor XIII B chain (1.09E+6 vs. 1.61E+6, p=0.020), apolipoprotein A-IV (2.30E+7 vs. 3.28E+7, p=0.024), alpha-enolase (1.02E+5 vs. 4.15E+4, p=0.024), heat shock protein 90-alpha (1.41E+4 vs. 4.08E+3, p=0.045), alpha-2-HS-glycoprotein (2.83E+7 vs. 3.68E+7, p=0.045), and neutrophil gelatinase-associated lipocalin (3.44E+4 vs. 1.36E+4, p=0.045). These results are summarized in the figure, which shows one group of proteins that decreased in abundance (on the left side of the volcano plot) and another that increased (on the right side) in the patients with low platelet function. The twelve proteins that changed the most fall into several categories related to platelet function. Low gelsolin and high histone levels are each associated with microvascular obstruction by release of intracellular material that accumulates in small vessels and activates platelets. Low HRG and high damage-associated molecular pattern (DAMP) protein levels are consistent with massive innate immune activation, which can impair platelet function in many ways.
Conclusion. This study provides an unbiased description of the change in proteomic profile associated with platelet dysfunction after trauma and identifies twelve proteins with the most profound changes. The pathways involving these proteins are salient targets for immediate investigation to better understand platelet dysfunction after trauma and identify targets for intervention.
Figure Legend. Volcano plot depicting fold-change and statistical significance of individual proteins in patients with low vs. high platelet function. Dotted horizontal line represents p = 0.05. Twelve proteins showed a statistically significant p value.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal